Research topics
- Combining live cell imaging and single-cell RNA sequencing
- Three-dimensional ultrafast live imaging of intracellular microstructure
- Large-scale automatized collection of data pairs of Raman Spectrum-RNA Sequence in a single cell
- Investigation of cell specificity responsible for MET events within kidney organoids using machine-learning approaches
- Nano fluidic chip -From micro to nano-
- Single cell analysis of human iPS-derived retinal organoid
- Exploration of a novel tissue stem cell population and niche
- Decoding the mechanism of neural epithelium formation during the development of neural organoids.
- Prediction of aging status using image data
- Integrated understanding of gene expression dynamics and cell differentiation lineages utilizing single-cell 3D genome information
- Development of a simple screening method based on the activity of molecules derived from a single gene
Combining live cell imaging and single-cell RNA sequencing
Summary
Recent technologies in next generation sequencing enable us to analyze whole genome expression in individual cells. However, analysis of whole genome expression in a cell basically destroys the cell. If we can predict the whole genome expression only by observing the cell, we can understand the details of internal cell state while keeping the cell alive. Toward this goal, we connect massive amounts of single-cell imaging data with single-cell genome expression data using machine learning.

Click to enlarge
Related labs
Three-dimensional ultrafast live imaging of intracellular microstructure
Summary
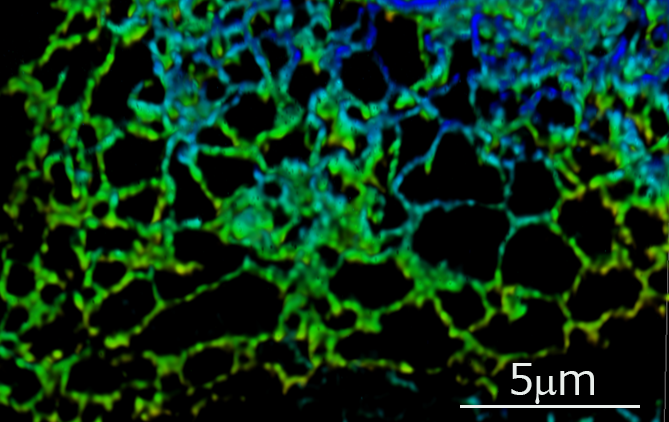
We observe the microstructure in the living cell with the world's highest shutter speed by using a uniquely developed "ultrafast super-resolution microscope". We DECODE the state of living cells noninvasively by detailed observation of morphology and dynamics of subcellular organelle such as mitochondria as well as microstructure in the nucleus.
Outer membrane of mitochondria captured by super-resolution microscope
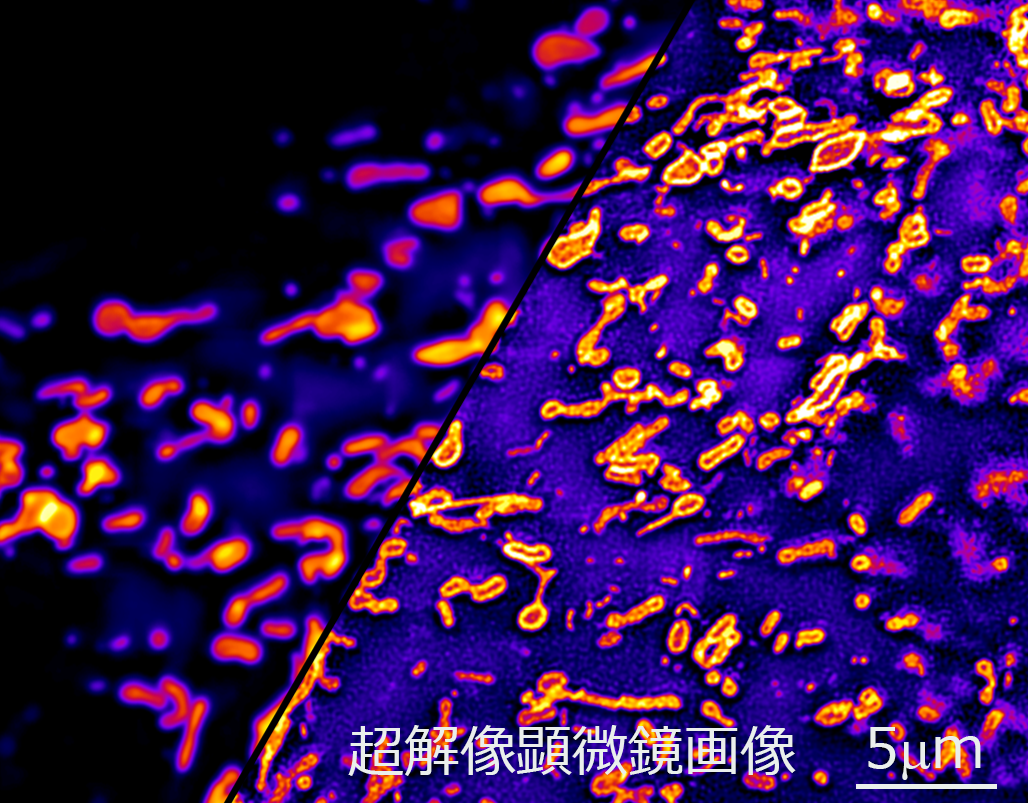
Super-resolution 3D live imaging of endoplasmic reticulum
Related laboratory
Large-scale automatized collection of data pairs of Raman Spectrum-RNA Sequence in a single cell
Summary
The aim of this project is to establish a technology for predicting gene expression from Raman scattering spectrum emitted from a cell so as to conceptively demonstrate how to "predict the intracellular information from scattering light". The Raman spectrum from a cell is determined by combinations of all molecular vibrational modes (CC, SH, benzene ring, etc.) inside the target cell. Since a cell is composed of various molecular species, the Raman spectrum will be too complicated to be decomposed. However, no matter how complex it is, if gene expression changes in the cell, the types and ratios of constituent molecules will change, and the Raman scattering spectrum is expected to change accordingly. In other words, the measured Raman spectrum is indirectly but surely correlated with the gene expression pattern via cellular function and/or state. From complexly modified Raman scattering light that can be measured non-invasively, it is possible to predict the gene expression that can only be measured invasively. To demonstrate the above concept, we collected data pairs of Raman spectrum and RNA sequence of 10 drug-resistant E. coli, and constructed a machine learning model to connect these two data pairs. With this machine learning model, we have succeeded in estimating the gene expression pattern of drug-resistant E. coli from the Raman spectrum with an accuracy of 80 to 90%. Furthermore, in order to demonstrate that the same concept holds for mammalian cells, we are carrying out the same experiment using disease-derived induced pluripotent stem cells (iPS cells). In addition, we are constructing a microscope system that enables automatic collection of Raman spectrum-RNA sequence data pair in a single cells.
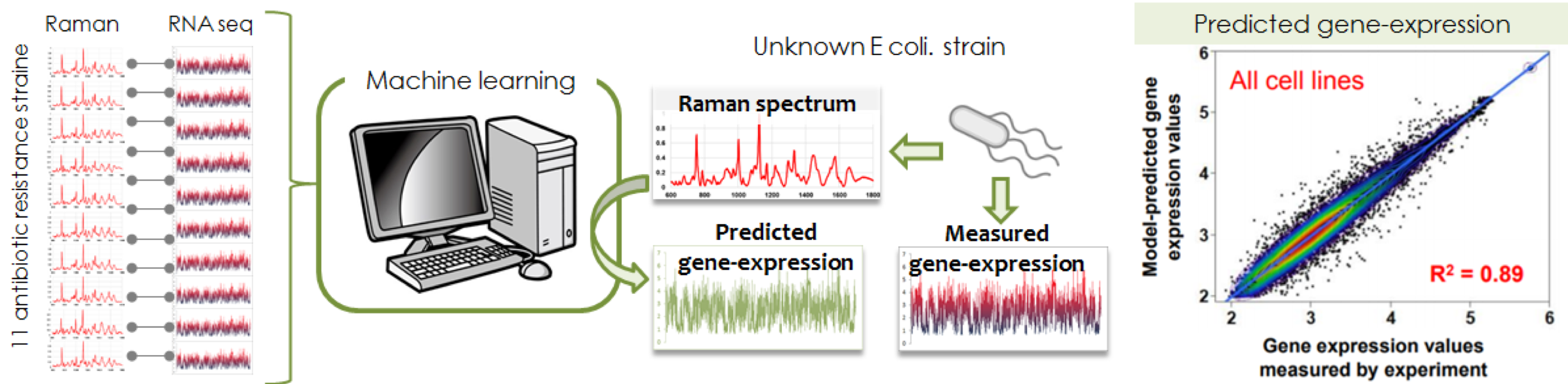
Click to enlarge
Investigation of cell specificity responsible for MET events within kidney organoids using machine-learning approaches
Summary
During kidney development, nephron progenitor cells develop into the nephron, a functional unit of the
kidney, via mesenchymal-epithelial transition (MET). We previously established an in vitro system in which
METs can be artificially induced to human iPSCs-derived nephron progenitor cells by the stimulation of
canonical-Wnt signalling. Interestingly, we further demonstrated that those MET events occur constantly
and singularly within a uniform nephron progenitor population.
Here, in this research project, we aim
to elucidate the mechanism that control such rare MET events using an in vitro MET occurrence system. For
this purpose, we will construct a system for real-time monitoring of MET events, and then use a machine
learning system to prospectively detect cells just before the onset of MET and analyze their
characteristics.
This study will help to elucidate the mechanism of determining the number of nephrons
in the human kidney, and the findings can be applied to the development of a new method for efficient
artificial induction of kidney tissue in the future.
Reference
- 1. *Takasato, M., Er, P.X., Chiu, H.S., and Little, M.H. (2016). Generation of kidney organoids from human pluripotent stem cells. Nature Protocols 11, 1681–1692.
- 2. *Takasato, M., Er, P.X., Chiu, H.S., Maier, B., Baillie, G.J., Ferguson, C., Parton, R.G., Wolvetang, E.J., Roost, M.S., Chuva de Sousa Lopes, S.M., et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568.
- 3. Takasato, M., and *Little, M.H. (2015). The origin of the mammalian kidney: implications for recreating the kidney in vitro. Development 142, 1937–1947.
Nano fluidic chip -From micro to nano-
Summary
We have many technologies on microdevice development for rapid and high-throughput analysis of cells and
has applied these technologies to life sciences. In BDR, we have fabricated and provided many microdevices
for cell and tissue analysis to many laboratories or projects. Recently, this concept has been extended to
molecules, namely nano scale and by combining with technologies for molecular measurement, simulation, or
analysis in BDR, we aim to pioneer a new scientific field.
However, nanoscale channels are not only
difficult to be fabricated, but also difficult to flow liquid in them. The scale is just between the
molecules and bulk. So, it was suggested that water and other molecules behaviors are different from bulk.
This field is difficult area not only as engineering but also science, but results with large impact is
expected.
The final goal of this project is to construct a platform to manipulate single molecules in
nanoscale. Before that, we aim to establish fundamental technologies including fabrication, fluid control,
and measurement method in nanochannel with Okada TL.
Related Labs
Single cell analysis of human iPS-derived retinal organoid
Summary
In November 2020, clinical research for the treatment of photoreceptor degeneration using neural retina sheets induced to differentiate from human iPS cells began. The neural retinal sheet used in this study was researched and developed at our institution, but at present, the retinal sheet for transplantation ismanually cut out fromtheneural retina toremove pigment epithelium and ciliary body.Butnot only does it require skillful techniques to cut out, but the size that can be cut out is as small as about 1 mm x 1mm, and many neural retinalsheets are required to cover a wide range of lesions, which requires enormous cost and labor. One solution is to develop a large neural retinal sheet. However, the technique for producing a largeretinal organoid that ensure qualityandsafetyhas not been establisheddue to the difficulties of controlling the differentiation from spatially and temporally heterogeneous cells.Therefore, in this project, we aim to develop a large-scale next-generation transplantation retinal sheet that suppresses differentiation into cells other than photoreceptor cells by reconstructing the retinal sheet only from retinal progenitor cells in a specific differentiation stage. Single cell (sc) RNA-seq is performed to understand the cell characteristics of retinal progenitor cells necessary for that purpose.
Exploration of a novel tissue stem cell population and niche
Summary
The tissue stem cells are the most important cell population in adult tissueshomeostasis. In particular,
the self-renewal and differentiation capacities of tissue stem cells are indispensable for tissue
metabolism and regeneration from damage. The airway epithelium is exposed to invasivebacteria, viruses,
and chemical stimulus thatmay damage the tissue. Airway epithelium has a relatively slow metabolism under
normal conditions, but upon injury, cells proliferate rapidly to repair the damaged area. This type of
tissue regeneration is called facultative regeneration in which stem cells activate their self-renewal
capacity in response to injury. Multiple tissue stem cell populationshave been identified in airway
epithelium that expected to respond to various type of damages to protect pulmonary system.
Recent innovations in stem cell culture techniques have made it possible to reconstitute mini-organs,
called organoids, by culturingadult tissue stem cells for long periods of time in the laboratory. In lung
organoid culture, lung tissue stem cells are isolated and grown into spheres by culturing them on a
three-dimensional scaffold, which is suitable for studying lung stem cell dynamics. In this research
project, we seek novel stem cells by combining scRNA-seq analysis with image analysis of lung tissue stem
cells and organoids, which will be performed in collaboration with the Dr. Shiroguchi’s Laboratory. The
candidates of the novel stem cell markers will be characterized using mouse lungs. In particular,in
vivostem cell properties will be verified based on cell dynamics during metabolism and injury
regeneration.
Decoding the mechanism of neural epithelium formation during the development of neural organoids.
Summary
Neural organoid is the three-dimensional neural tissue that is derived from pluripotent stem cells (PSCs). It is elusive how the neural epithelium develops during self-organizing formation of neural organoids. In this research subject, we will first induce neural organoids using knock-in human PSCs that can determine the generation of neural epithelium, and then perform 4D imaging to capture how the epithelial structure is constructed. Finally, we will elucidate the genetic background of the self-organizing formation of epithelial structure by single-cell RNA-seq analysis at the key point of the process. Combining the information of imaging and RNA-seq, it will be possible to explain how the complexed epithelial structure emerge.
Prediction of aging status using image data
Summary
Aging is a biological process that occurs over an entire lifetime. Because aging is a rather complex process that takes a long time and is sensitive to genetic and environmental factors, a comprehensive understanding of the aging process remains unsolved. This research plan aims to comprehensively understand the aging process by monitoring changes in appearance, gene expression, metabolism, and muscle function over an entire lifetime using a genetically homogeneous model organism C.elegans. Furthermore, we examine the links between obtained data using the machine learning approach. This study would help us deepen our knowledge of the aging process and might predict the state of aging of an individual animal based on its appearance.
Integrated understanding of gene expression dynamics and cell differentiation lineages utilizing single-cell 3D genome information
Summary
The three-dimensional organization of the genome within the cell nucleus is complex and plays an important role in various genome activities, including the regulation of transcription for gene expression. However, many aspects of the relationship between this 3D genome and gene regulation remain unelucidated. This project aims to deeply understand the relationship between the 3D genome structure and gene expression dynamics by integrative analysis using omics technologies. Furthermore, we aim to develop single-cell multi-omics technologies that simultaneously acquire 3D genome and gene expression. Through this study, if we can understand cellular states from a new perspective utilizing the information of the 3D genome structure, which differs from gene expression information, it is expected that this will lead to the development of technology for predicting cell differentiation lineages in the future, utilizing machine learning and other advanced techniques based on that information.
Development of a simple screening method based on the activity of molecules derived from a single gene
Summary
When conducting genetic functional analysis of biopolymers or aiming to obtain molecules with desired properties and functions, evolutionary molecular engineering techniques are extremely useful because it can screen target molecules from a vast library of molecules by reproducing "mutation → selection → amplification" in vitro. However, typical evolutionary molecular engineering often selects molecules based on binding affinity. Therefore, we need to devise methods for selecting molecules focusing on functions beyond binding affinity. In this research, we aim to develop a simple method for activity-dependently selecting molecular variants expressed in artificial cells, intended as a general-purpose system. The system is composed of 3 steps as follows. First, mutant genes are individually encapsulated into semi-permeable membrane vesicles that allow only small molecules of about 2 to 3 kDa or smaller to pass through. Next, mutant genes are transcribed and translated into the target molecule in each vesicle using the cell-free protein synthesis system, the PURE system [Shimizu Y, Nat. Biotechnol., 2001]. Finally, fluorescent, chromogenic, and luminescent substrates infiltrate from outside the vesicle, and vesicles containing mutants are screened based on the light intensity when these substrates react with the molecule. This method is expected to enable us to obtain molecules with desired functions from a library of over approximately 105. If this research is successful, it will be possible to quantitatively measure the activity of biopolymers under various physiological conditions, such as the type and concentration of salts and metal ions, and pH. This will enable the analysis of the functions of target molecules in a general-purpose manner. It is also expected to contribute to the acquisition of enzymes that exhibit higher activity than natural molecules, and to the development of novel probes, biosensors, and drug candidate molecules.
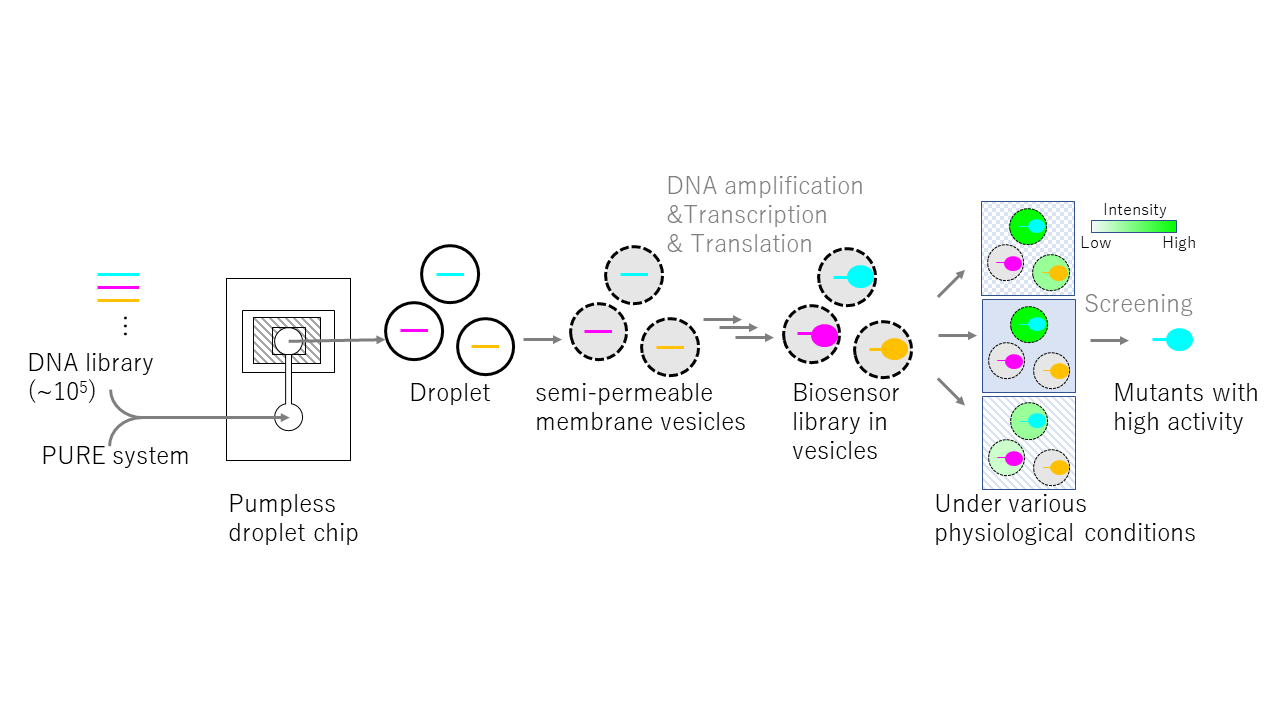
Click to enlarge